Kinetic Modeling based Probabilistic Segmentation for Molecular Images
Abstract
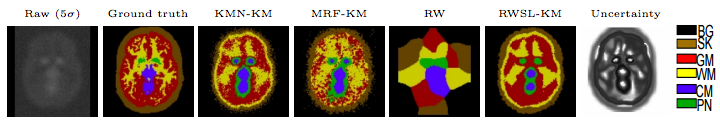
We propose a semi-supervised, kinetic modeling based segmentation technique for molecular imaging applications. It is an iterative, self-learning algorithm based on uncertainty principles, designed to alleviate low signal-to-noise ratio (SNR) and partial volume effect (PVE) problems. Synthetic fluorodeoxyglucose (FDG) and simulated Raclopride dynamic positron emission tomography (dPET) brain images with excessive noise levels are used to validate our algorithm. We show, qualitatively and quantitatively, that our algorithm outperforms state-of-the-art techniques in identifying different functional regions and recovering the kinetic parameters.
 Top
Top
- Möller, Torsten
- Saad, Ahmed
- Hamarneh, Ghassan
- Smith, Ben
 Top
Top
Supplemental Material
Shortfacts
Category |
Paper in Conference Proceedings or in Workshop Proceedings (Full Paper in Proceedings) |
Event Title |
Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008 |
Divisions |
Visualization and Data Analysis |
Event Location |
New York |
Event Type |
Conference |
Event Dates |
Sep. 6 - 10, 2008 |
Publisher |
Springer |
Date |
2008 |
Export |
 Top
Top




